Overview
Having established a world-class research environment with wide-ranging applications and technology platforms (Flow Cytometry, Next Generation Sequencing, Protein Expression, Advanced Microscopy & Imaging, High-Performance Computing, and Bioinformatics & Data Management), the Centre is now well-positioned to conduct cutting-edge research into an expanded range of diseases. The Centre’s projects now feature a larger research portfolio, with its integration of human genetics to facilitate research into host-pathogens interactions, while serving as a bridge between IDs and NCDs.
To effectively support research and training at the Centre, and ensure efficient use and longevity of the equipment, we have organized the facilities into six technology platforms, each led by a scientist with specific expertise to manage and proceed user training for the facilities. These include:
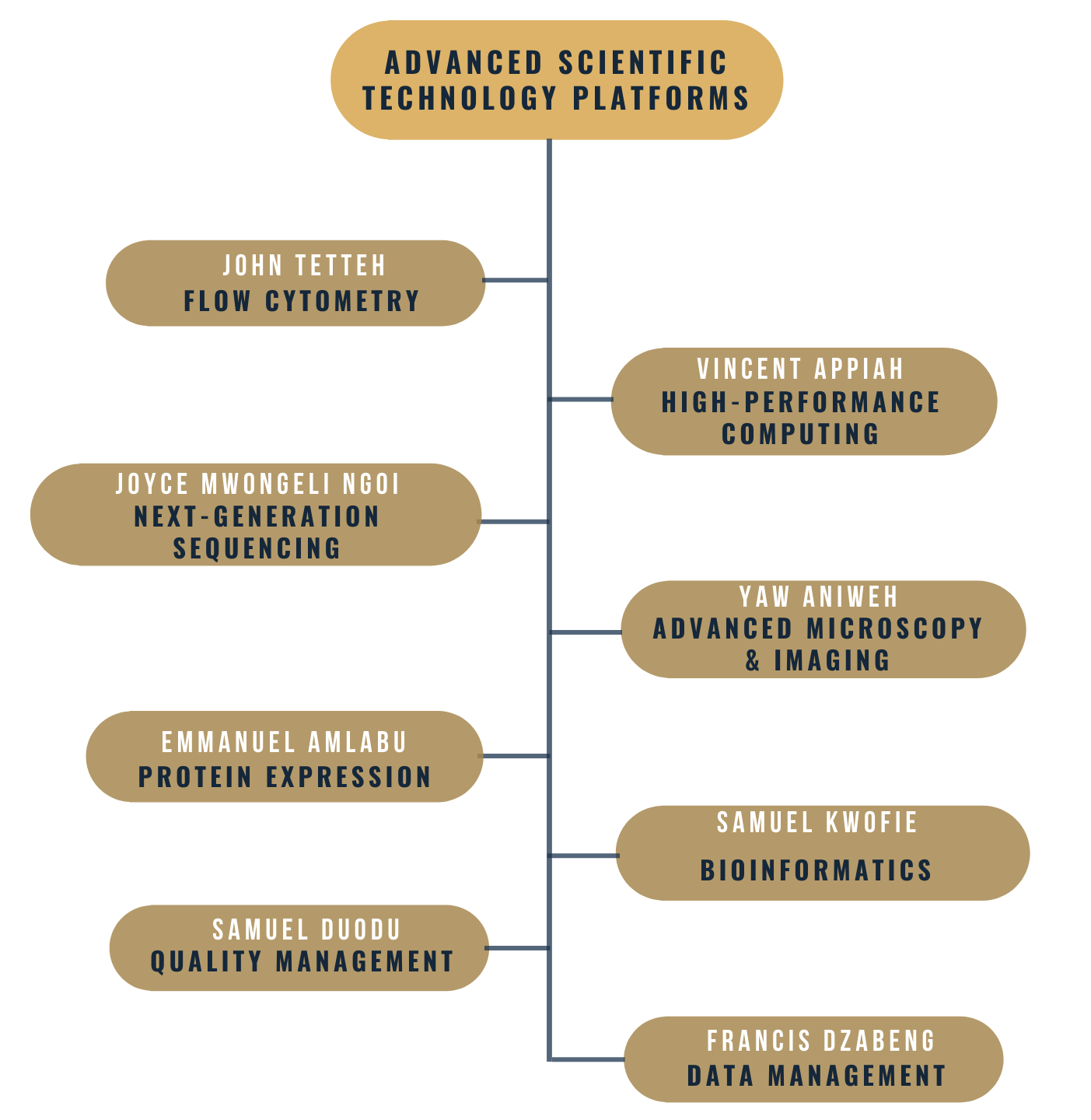
- Flow cytometry (FACS), including fluorescent activated cell sorting and phenotypic and functional analyses of cells;
- Next generation sequencing (NGS), for genomic and transcriptomic analysis of host and pathogen DNA/RNA;
- High-performance computing (HPC), for providing storage and computational power to support analysis of genomic, transcriptomic, and proteomic data;
- Advanced microscopy and imaging (AMI), for fluorescent and confocal microscopy to support cell biology and functional characterization of proteins;
- Protein expression (PE), for cloning and expression of drug and vaccine candidate genes and production of enzymes for downstream applications; and
- Bioinformatics and data management (BDM), which provides high-specification computers with various applications and statistical software for analysis of clinical, genomic, and transcriptomic data.
Priority Diseases
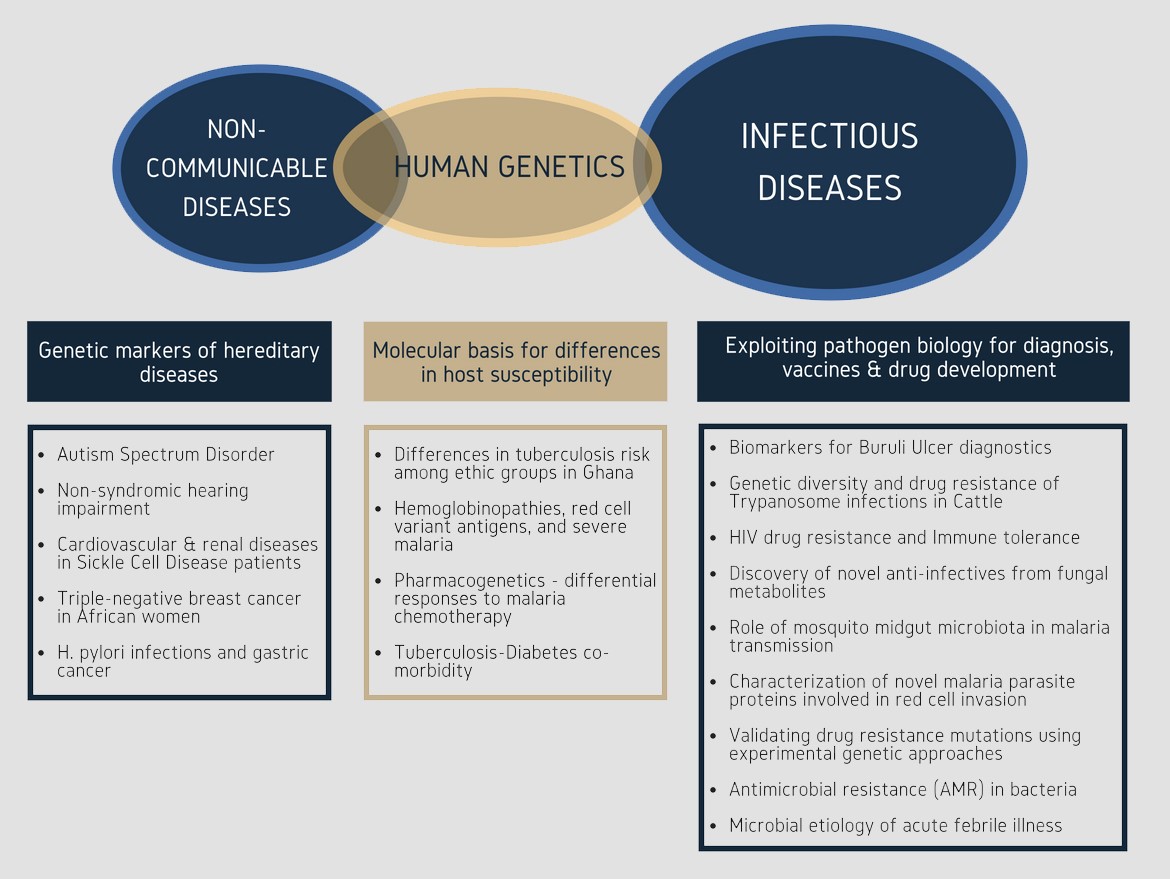
- biomarkers and molecular diagnosis
- disease pathogenesis and immunity;
- pathogen and host genomics and host-pathogen interactions;
- molecular epidemiology for disease surveillance and drug resistance monitoring; and
- target discovery for drug and vaccine development.
Research Focus
IDs currently contribute the highest proportion of total disability adjusted life years (DALYs) lost in sub-Saharan Africa (sSA), with the majority of mortality in sSA attributed to malaria, tuberculosis, bacterial infections, and human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS). However, by the year 2030, four NCDs, namely cardiovascular disease, cancer, respiratory diseases and diabetes, are projected to be the primary cause of DALYs lost in sSA. This presents a daunting challenge for many countries in sSA, where scarce resources and inadequate health care infrastructure severely limit the capacity to respond effectively to the double burden of disease. While presenting a significant public health concern, co-morbidities of IDs and NCDs provide a unique opportunity to understand the relationship between infectious exposures and NCDs in ways that are no longer possible in resource-rich nations.
- Exploiting pathogen biology for developing novel disease diagnostics, vaccines and drugs;
- Determining the molecular bases for differences in host susceptibility to IDs to guide better disease prevention and management;
- Identifying genetic markers to inform molecular diagnostic approaches for early detection of NCDs.
These objectives are embedded within the core aims of specialised research groups that work co-dependently, covering several disease research areas that fall under our braoder research themes. The research groups contibute world-class context-specific research that focuses on finding solutions to the many healthcare challenges in Ghana and across the African continent.

The malaria group is the largest at the Centre, and it is led by Profs Gordon Awandare and Neils Ben Quashie. They employ an immunology-driven approach aimed at deconstructing the mechanisms of natural immunity to malaria and how they could be exploited for vaccine development. The group uses a systems immunology approach, which has recently been perfected through ongoing research with our collaborators at the Francis Crick Institute in the United Kindom. This involves incorporating cellular, transcriptomic, cytokine and antibody data to determine immune signatures associated with natural immunity, using a longitudinal design that will enable comparison of the kinetics and composition of immune responses over the course of an acute P. falciparum infection in children with different levels of pre-existing immunity. A similar approach will be used to study placental malaria, which is a major threat to maternal and neonatal health. The Antigen discovery approach of the malaria group uses a combination of informatic and functional analyses of Plasmodium antigens from both asexual and sexual stages of development, with the goal of identifying novel proteins that could be targeted for vaccines or drugs.
The Genetics and Genomics approach is based on parasite population genetics, studying genomic variation of parasites circulating in endemic areas and identifying antigens that may be under selective pressures. Genes of interest would be evaluated by genetic manipulations. From the host perspective, the role of genetic variation in immune response mediators and red blood cell antigens will be investigated to determine the mechanisms of natural resistance to malaria.
The Malaria group has an extensive network of collaborators both regionally and internationally, including Dr. Alfred Ngwa (MRC The Gambia), Prof Mahamadou Diakite (Malaria Research and Training Centre, Mali), Prof David Conway (London School of Hygiene and Tropical Medicine), Dr. Jean Langhorne (Francis Crick Institute, London), and the MalariaGen consortium, led by Prof Dominic Kwiatkowski (Wellcome Trust Sanger Institute, UK).
The Molecular Virology Research group is led by Dr. Osbourne Quaye and conducts research on various viruses including gastro-viral agents, human immunodeficiency virus (HIV), Hepatitis B and C, and filoviruses among others. Studies investigate the efficacy of the existing rotavirus vaccine in unsanitary environments and the contribution of other gastro-viral agents in these environments.
Additional novel projects investigate the immune checkpoint molecules that are activated in HIV infection in long-term non-progressors to determine the mechanisms of natural protection and how they can be exploited for treatment and vaccine development. The group also ivestigates mechanisms of drug resistance in HIV patients, with particular attention to the elucidation of mechanisms of resistance to Integrase inhibitors in HIV strains that predominate in West Africa. The group is also working on developing and validating assays that can routinely be used to screen animal species (including bats) implicated to cause zoonotic filovirus infections such as Ebola for exposure to these viruses.
WACCBIP and its partners have been at the forefront of the response to the COVID-19 pandemic in Africa. For example, in Ghana, we have contributed to testing of suspected cases, undertaken genomic sequencing of viral samples, and created a repository of blood samples from diagnosed patients. Our research goals for COVID-19 focuses on examining the contributions of viral genomic diversity, and host innate and adaptive immune responses, in determining the clinical course of the disease in Africa.
The Virology group has established strong collaborations with colleagues at the University of Cambridge, including Profs James Wood and Olivier Restif from the Department of Veterinary Medicine and Dr. Edward Wright from the University of Sussex.The Microbial Pathogenesis and AMR group is led by Prof Dorothy Yeboah-Manu and Dr. Lydia Mosi. The group works on a range of bacterial pathogens, including Mycobacteria that cause tuberculosis and Buruli ulcer, food and water borne pathogens such as Salmonella typhi, Vibrio cholerae and Escherichia coli that cause frequent outbreaks and respiratory pathogens such as Klebsiella pneumoniae.
There is also specific interest in hospital acquired infections, especially bacteria of the genus Enterobacter, which are known to thrive in hospital environments. For all these pathogens, the investigations focus on genetic diversity and virulence factors, which are critical for disease surveillance. In addition, the threat of AMR is being addressed, where genetic markers of antibiotic resistance as well as the role of mobile genetic elements in the spread of resistance are under investigation.
This group has major strategic collaborations with Profs Miguel Valvano (Queen’s University, Belfast), and Sebastian Gagneux (Swiss Tropical Public Health Institute).
This group is led by Dr. Theresa Manful Gwira, and focuses on non-plasmodium protozoan diseases, specifically trypanosomiasis, leishmaniasis and babesiosis. The research aims to characterize these infections, including in livestock, to identify certain molecular, biochemical and immunological signatures which could improve diagnosis of the diseases. The group also seeks to explore all possible drug resistance mechanisms of trypanosomes against the existing therapeutic and prophylactic drugs used in treatment and identify novel immunogenic proteins that can be used as possible vaccine candidates.
This group collaborates with some of the leading experts in protozoan cell biology, including Prof Keith Gull (Oxford University) and Prof Mark Carrington (University of Cambridge).
This group is led by Dr. Patrick K. Arthur and its research is towards the discovery and development of new antibiotics against tropical infectious diseases using fungal metabolites. The group is using a chemical systems biology approach and has so far isolated and screened more than 300 distinct fungal species, for which there was a high infective discovery rate.
A phenotypic array-based workflow has now been developed which allows the comparison of the bioactive extracts to many standard antibiotics based on their pattern of activities across different physiological conditions of the test organisms. Going forward, pure compounds will be isolated from fungal extracts from the priority list and extensively characterized using NMR-structural elucidation, advanced imaging techniques and mass spectrometry-based proteomics to characterize their mechanisms of action and pave the way for pre-clinical development.
This group is led by Prof Regina Appiah-Opong and is working on investigating genetic and cellular mechanisms for common cancers and the roles of IDs in oncogenesis in cancers such as gastric, liver, cervical etc. The research also includes investigation of mechanisms of chemo-resistance mediated by a sub-population of cells called cancer stem cells (CSC). Specific studies will be geared towards identifying genetic risk factors for triple negative breast cancer and new drug targets for prostate cancer. Identification of compounds that affect CSC activity and the molecular mechanisms involved will also be investigated, including the role of interleukin-8.
A phenotypic array-based workflow has now been developed which allows the comparison of the bioactive extracts to many standard antibiotics based on their pattern of activities across different physiological conditions of the test organisms. Going forward, pure compounds will be isolated from fungal extracts from the priority list and extensively characterized using NMR-structural elucidation, advanced imaging techniques and mass spectrometry-based proteomics to characterize their mechanisms of action and pave the way for pre-clinical development.
This group will be led by Prof Augustine Ocloo, who has been leading efforts towards building capacity in the sub-region for the early detection and management of mitochondrial disorders. They are employing an innovative approach based on the role of mitochondrial dysfunction as a mechanism for some common illnesses and chronic conditions, such as diabetes and heart disease, which are due to genetic defects that reduce the ability of the mitochondria to produce energy. Although difficult to diagnose, recent advances in mitochondrial physiology research demonstrate that examination of mitochondrial oxidative capacity in tissue biopsy in combination with blood, urine and mtDNA mutation analyses could provide strong evidence for early detection and management of these disorders.
This research group is led by Dr. Prosper Kanyong and is focused on developing innovative biosensor research and technology for use within resource-limited settings. The group aims to develop and apply nanomaterials and devices for both the detection of several specific disease biomarkers, as well as general chemical/biological mediators of relevance for disease diagnostics, environmental monitoring and food analysis.
Research Engagements
WACCBIP is keen to strengthen its evolving partnership and engagements with local public sector organizations such as the National Malaria Control Programme (NMCP), The National Buruli ulcer Control Programme (NBUCP), and the National TB Control Programme, and key national and international industrial sector partners such as the Pharmaceutical Society of Ghana (PSGH), Novartis Institute for Tropical Diseases (NITD), the Centre for Proteomics and Genomics Research (CPGR), Cape Town, and Inqaba Biotechnical Industries ltd, Pretoria, South Africa.





